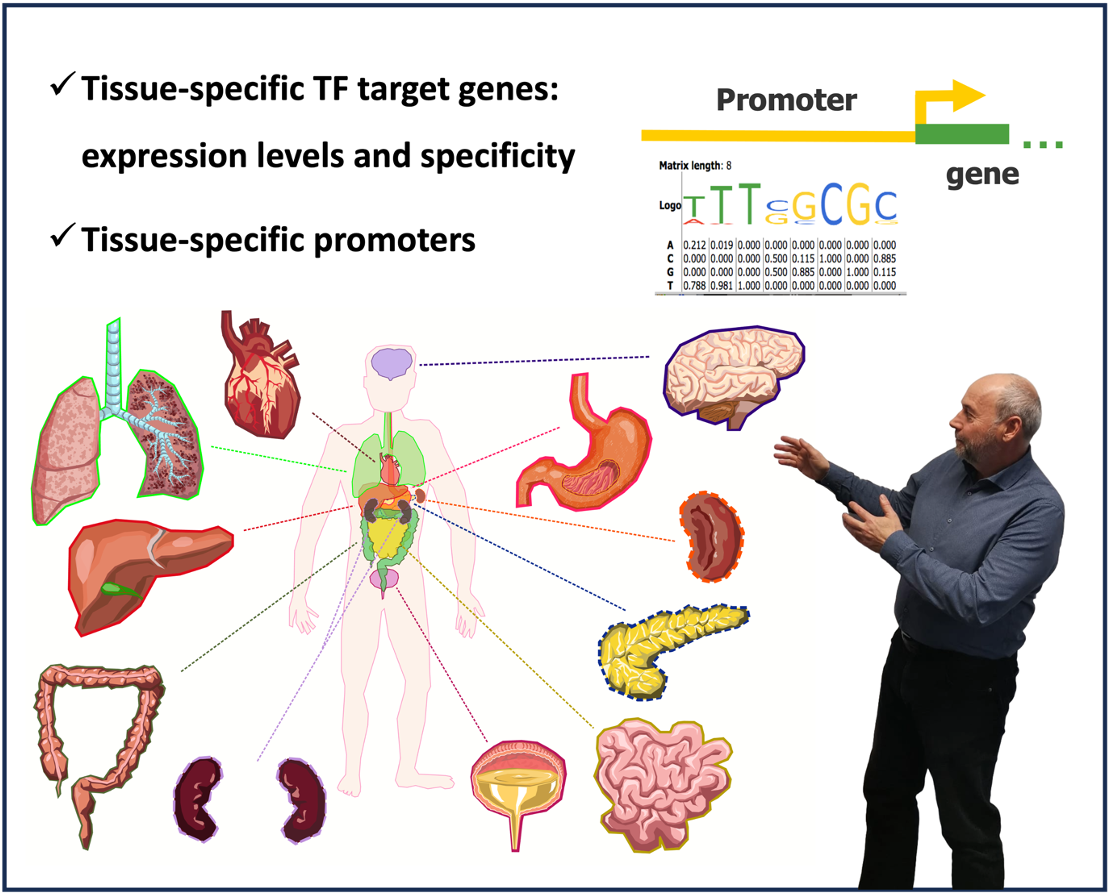
How to find tissue-specific transcription factor (TF) target genes?
Q&A: Coffee break with TRANSFAC
This event was held on May 21st, 2024
FREE online seminar
Seminar Description
In this webinar Prof. Dr. Alexander Kel will share with you the latest intriguing insights of gene regulation mechanisms. He will demonstrate how to perform tissue-specific gene regulation analysis and find enriched motifs using the TRANSFAC database and its MATCH Suite tool.
The following main questions will be in the focus of this seminar:
Seminar Schedule
The session started on May 21 with a lecture from Prof. Dr. Alexander Kel (approx. 1 hour) and was continued with a Q&A session addressing the questions coming from the attendees of the online event (up to 30 minutes).
What You Get
The Lecturer

Prerequisites
Basic knowledge of biochemistry and molecular biology. No programming skills are required.